Dear readers,
Just two days after the release of our previous newsletter on BMS’ new schizophrenia drug, Cobenfy, Time named it to be one of the top 100 inventions of 2024. The full list is linked here for those searching for rabbit holes to go down. Today’s edition will be a pivot from schizophrenia and FDA approvals to a topic that probably occupied the other 99 spots on that Time list – AI. Specifically, I want to introduce one company that is working on some cool science and talk about some of the methods that they’re using, which are becoming more common in today’s age of AI.
In the summer of 2023, layoffs at Meta impacted a team of researchers working to understand how AI models could be used to design proteins. Immediately, Alex Rives, who led the team at Meta, spun up a new startup based on the work they had been doing. In June of 2024, EvolutionaryScale announced that they raised $142M, with support coming from Amazon, Nvidia, Lux Capital, among others.
Using advanced computational methods to understand proteins has recently been in the news quite a bit, with the Nobel prize in chemistry being awarded to Demis Hassabis and John Jumper of DeepMind for their work in developing AlphaFold (covered in a previous edition of this newsletter) and David Baker from the University of Washington, for his work in computational protein design. EvolutionaryScale’s AI model is called ESM3 and is an impressive step in an already big year for protein design. Their mission is “to develop artificial intelligence to understand biology for the benefit of human health and society, …”. If you visit the website of any company working at the intersection of AI and biology, you’ll likely find something along those lines. So what is EvolutionaryScale actually working on here?
ESM3 falls under a broad class of models called generative AI models, which also includes OpenAI’s ChatGPT, Google’s Gemini, Anthropic’s Claude, among many others. The idea behind these models is that if you give them enough data, they will eventually be able to learn and mimic patterns that are found in the data. Let’s take the example of the massively popular large language models for text. At a very high level, what is going on is that these models take in huge amounts of data from all over the internet and break them down into what are called “tokens”. In the case of these chatbot language models, a token can be thought of as an individual word (though it may sometimes be just part of a word). The model is trained to learn connections between the tokens, or words. For example, it may learn that the token “peanut” often appears before the token “butter”, but less often before the token “isthmus”. Part of the process of building these connections is known as “masking”, where some of the tokens in the data are hidden and the model tries to fill in the blanks according to the context. With each attempt to fill in the blanks, the model tweaks its internal settings, called “weights”, to better understand relationships between tokens. By repeating this process many times across huge datasets, the model gradually becomes better at predicting the next token.
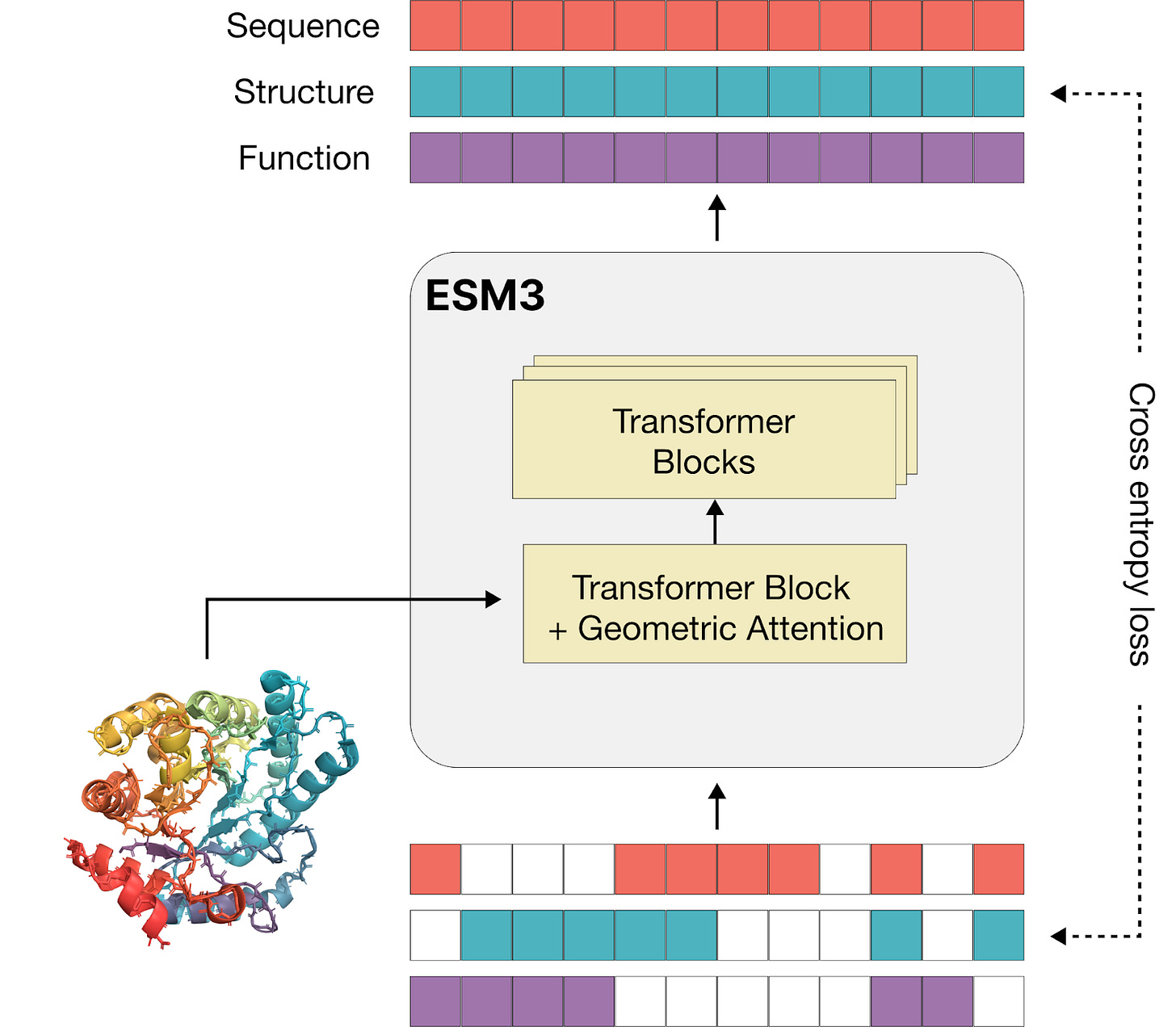
So what about in the case of ESM3? Well in this case, Rives and his team wanted to design a model that could express the sequence, structure, and function of proteins. To do this, they created a custom token structure that captures all of this information. In order to train ESM3, they took data from billions of proteins and encoded them in this custom token structure. They were then able to use the masking strategy to learn connections between the sequence, structure, and function of proteins. I included a diagram below from the preprint describing the ESM3 model. If it doesn’t make a ton of sense, ignore it and forget that I ever brought it up.
In the preprint, the EvolutionaryScale team presents a demo of how powerful the ESM3 model can be using green fluorescent proteins, or GFPs. GFPs are a naturally occurring protein that are able to emit green light. They are an extremely useful tool in biotechnology because of their natural fluorescence, range of variants, and versatility in experiments. When given the partial structure of a GFP, ESM3 was able to generate other candidates for GFPs. The team then took one of the more promising options, giving ESM3 its partial structure in order to generate more new candidates. Of these, they found that one of the suggested new proteins (referred to in the preprint as esmGFP) achieved a similar degree of fluorescence to the most similar naturally occurring GFP, with just 58% sequence similarity. The authors compare the process of completing the missing segments of protein structure to how evolution navigates protein design in nature. Using a simulation tool to judge evolutionary distance between the newly generated esmGFP and the most similar naturally occurring GFP, they find that the distance represents 500 million years of natural evolution.
Like ESM3, previous versions of the model have been released to the public so that researchers can use it for their own projects. One example of how this has been used came out of Stanford University, when researchers used an older ESM model to generate new antibodies with improved binding stability and viral neutralization, to make them more appealing for therapeutic development.
With brevity in mind, I have left some information out of this note. I will leave the more technically-inclined readers with this tweet by Pranam Chatterjee, an Assistant Professor at Duke University, who gives an academic’s view on the pros and cons of ESM3. And as always, for those interested, please see the referenced literature below to help guide your journey in learning more.
Have a rejuvenating and joyful week!
Tommer
Thanks for reading & subscribing to Compbio, Pharma, and Ramblings! If you found this newsletter informative, it’d mean the world if you shared it with others 👩🔬
Referenced Literature:
Nature: Ex-Meta scientists debut gigantic AI protein design model
Biorxiv: Simulating 500 million years of evolution with a single language model
Nature: Chemistry Nobel goes to developers of AlphaFold AI that predicts protein structures
Nature: Efficient evolution of human antibodies from general protein language models